Archives
- 2026-02
- 2026-01
- 2025-12
- 2025-11
- 2025-10
- 2025-09
- 2025-04
- 2025-03
- 2025-02
- 2025-01
- 2024-12
- 2024-11
- 2024-10
- 2024-09
- 2024-08
- 2024-07
- 2024-06
- 2024-05
- 2024-04
- 2024-03
- 2024-02
- 2024-01
- 2023-12
- 2023-11
- 2023-10
- 2023-09
- 2023-08
- 2023-06
- 2023-05
- 2023-04
- 2023-03
- 2023-02
- 2023-01
- 2022-12
- 2022-11
- 2022-10
- 2022-09
- 2022-08
- 2022-07
- 2022-06
- 2022-05
- 2022-04
- 2022-03
- 2022-02
- 2022-01
- 2021-12
- 2021-11
- 2021-10
- 2021-09
- 2021-08
- 2021-07
- 2021-06
- 2021-05
- 2021-04
- 2021-03
- 2021-02
- 2021-01
- 2020-12
- 2020-11
- 2020-10
- 2020-09
- 2020-08
- 2020-07
- 2020-06
- 2020-05
- 2020-04
- 2020-03
- 2020-02
- 2020-01
- 2019-12
- 2019-11
- 2019-10
- 2019-09
- 2019-08
- 2019-07
- 2019-06
- 2019-05
- 2019-04
- 2018-07
-
Based on the above we hypothesized that a PROTAC strategy
2020-08-20
Based on the above, we hypothesized that a PROTAC strategy would be effective to induce CK2 protein degradation. Herein reported is an approach for the preparation of novel PROTACs via “click reaction” for degradation of CK2 protein (Fig. 2). Importantly, “click reaction” is a very facile, selective
-
br Introduction Myotonic dystrophy dystrophia myotonica DM i
2020-08-20
Introduction Myotonic dystrophy (dystrophia myotonica, DM) is an autosomal dominant disorder and the most common form of inherited muscular dystrophy in adults [1]. DM is characterised by a wide range of symptoms, including myotonia, progressive muscle loss, cataracts, cardiac conduction defects,
-
Our recent QTL mapping study revealed that
2020-08-20
Our recent QTL mapping study revealed that the adaptor genes, including the NCK genes and the ABI genes, are located within the genomic regions harboring the most significant QTL (Zhou et al., 2017), suggesting that these genes could be involved in the determination of resistance, and possibly throu
-
br Acknowledgments We thank Jon
2020-08-19
Acknowledgments We thank Jon Clardy, Juan Manuel Domínguez, José Francisco García Bustos, and the GlaxoSmithKline (GSK) Plasmodium falciparum dihydroorotate dehydrogenase project team for their contributions, helpful advice, and discussions. Introduction Dihydroorotate dehydrogenase (DHODH) i
-
br Four active site residues of KSTD are fully
2020-08-19
Four active site residues of Δ1-KSTD1 are fully conserved in Δ1-KSTDs from different species (Supplementary Figure S2). These residues are Tyr-119, Tyr-487, and Gly-491 from the FAD-binding domain and Tyr-318 from the catalytic domain. The structure of the Δ1-KSTD1•ADD complex revealed that the hy
-
WZB117 Though limited to a large cohort of self reported
2020-08-19

Though limited to a large cohort of self-reported healthy individuals, associations between individual WZB117 have been identified which may be clinically significant. Though slight, there is a relationship between the CYP2D6-inferred metabolizer phenotype and the diplotype-predicted activities of
-
In addition to providing substantial insight into substrate
2020-08-19
In addition to providing substantial insight into substrate recognition, structural studies have revealed important details about mechanisms of glycosylation and HCF-1 cleavage [28,30,33]. Activation of the nucleotide-sugar is accomplished through interactions of the β-phosphate with the N-terminus
-
br Results br Discussion Our UbV
2020-08-18
Results Discussion Our UbV library was originally designed to develop inhibitors of deubiquitinases (Ernst et al., 2013). Recently, we showed that UbVs could exhibit multiple binding modes and mechanisms to modulate HECT E3 activity (Zhang et al., 2016)—one set occupied the HECT domain E2-bind
-
The E enzyme is the
2020-08-18
The E1 enzyme is the apex for downstream enzymatic cascades and signaling pathways mediated by Ub and Ub-like proteins (Ubls) (Pickart, 2001). Studying characteristics of E1 and its catalytic functions may throw light to the role of ubiquitination in cell development. All known eukaryotic E1 are mon
-
If the unwanted objects detected by OpenComet are manually d
2020-08-18
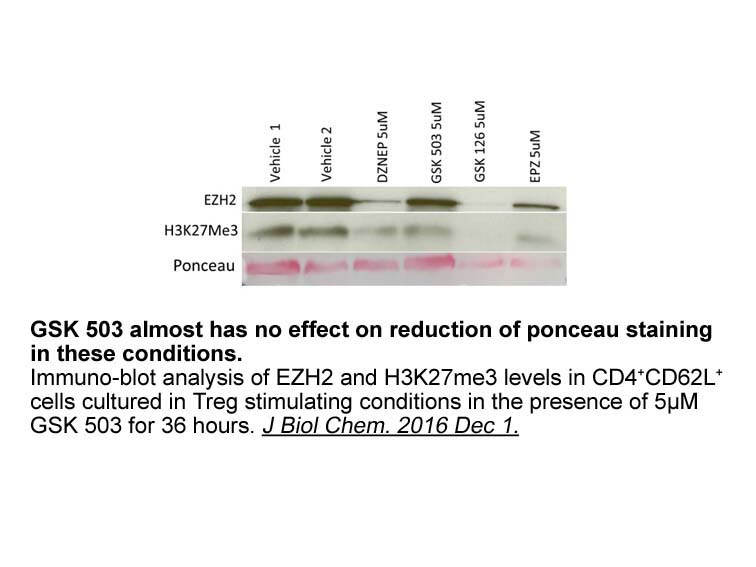
If the unwanted objects detected by OpenComet are manually deleted, and then the DNA (%) in tail is computed, the performance of the present method can be compared more effectively. Therefore, performance of OpenComet is analysed in two ways as Approach 1 and Approach 2. Approach 1, considers the DN
-
Our results indicate a deregulation of DNA methylation
2020-08-18
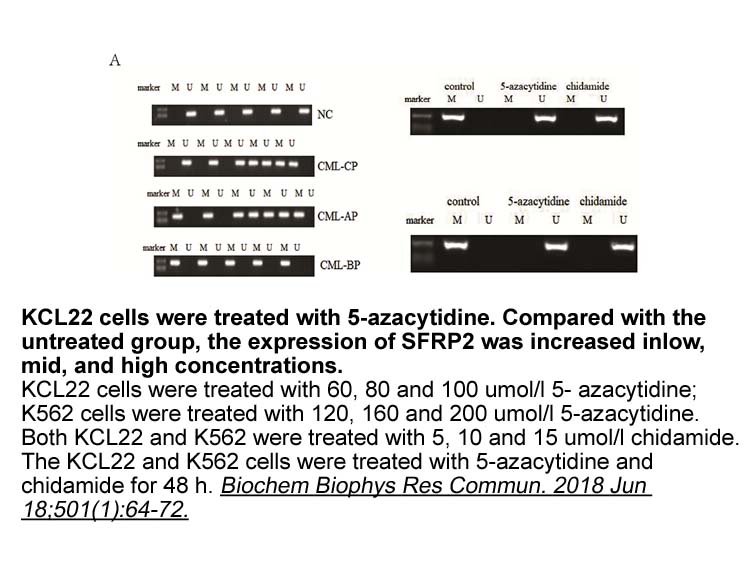
Our results indicate a deregulation of DNA methylation machinery during seed development in apomixis. Since DRM2 is targeted loci through an RNA-directed DNA methylation (RdDM) pathway involving 24-nt siRNAs (Stroud et al., 2014, Law and Jacobsen, 2010) and differentially expressed between apomict a
-
Collectively the results presented here provide new insights
2020-08-18
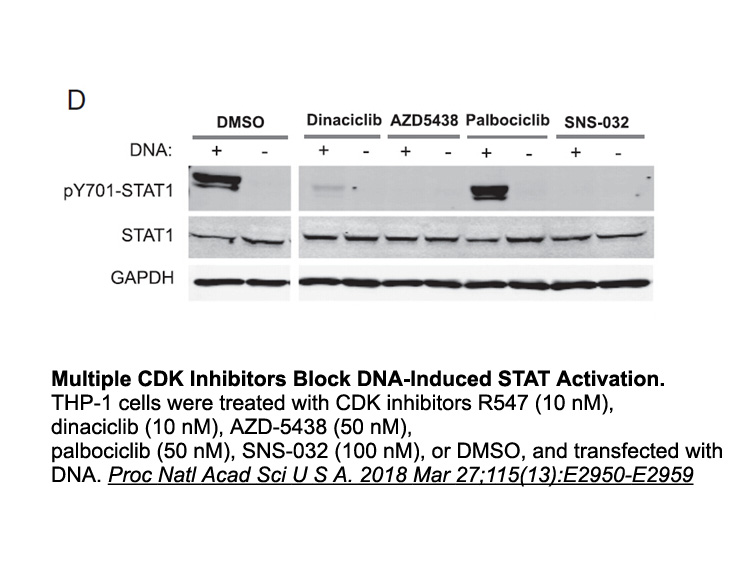
Collectively, the results presented here provide new insights into ligand binding, clustering, spatial distribution, and phosphorylation of DDR1b and DDR2 in response to soluble collagen I. As depicted in the cartoon of Scheme 1, we postulate a model in which the spatial distribution and assembly of
-
We found that both DDR and DDR
2020-08-18
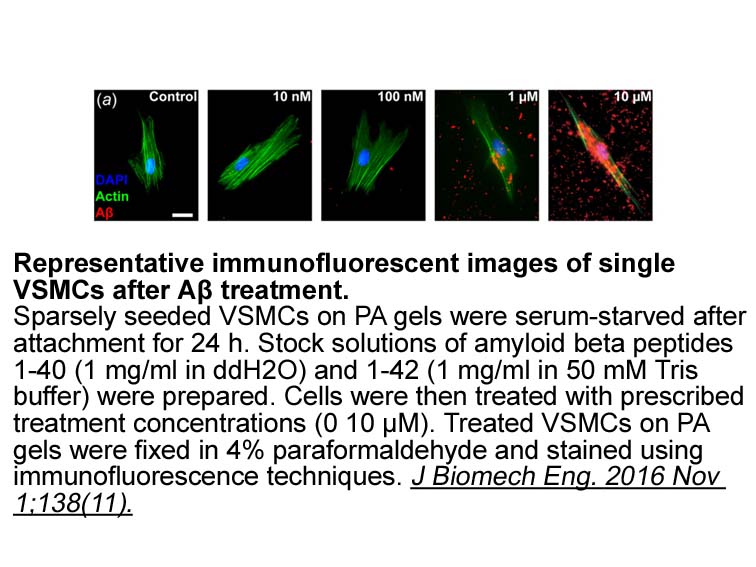
We found that both DDR1 and DDR2 ECD increased matrix mineralization as compared to native cells, with the effect of DDR2 ECD being more prominent. Both soluble (DDR2/ECD) and membrane-bound DDR2 ECD (DDR2/-KD), when compared to wild-type cells, induced larger mineral deposits. In this regard, a rec
-
Triclosan The annotation is arguably the most important part
2020-08-18
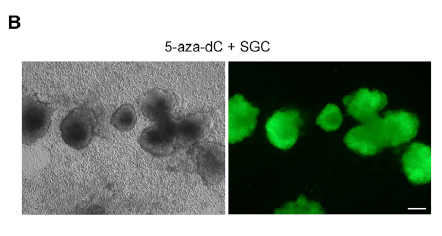
The annotation is arguably the most important part of analysis, as it enables one to evaluate and interpret the content of the transcriptome assembly. In this context, the non-redundant contigs were initially run in BLAST against the Nr database showing that 55% of them displayed high similarity to
-
As suggested by previous data we identified
2020-08-18
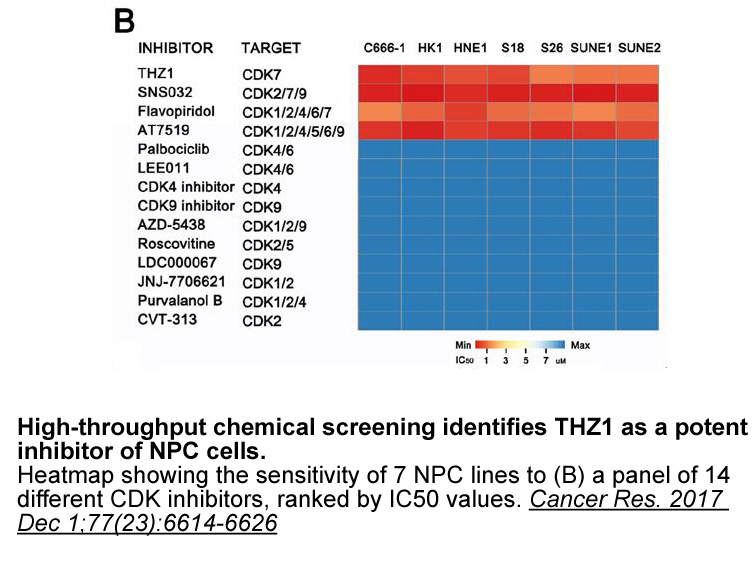
As suggested by previous data [45], we identified that the N-terminal and C-terminal domains of DDX3 were enriched in intrinsically disordered regions (Fig. 6). We also found that this characteristic was conserved in the external domains of several homologs of DDX3 (from yeast to human) and all desc
15578 records 787/1039 page Previous Next First page 上5页 786787788789790 下5页 Last page